For this tutorial, download and open this tulip graph file. This graph represent relations between authors, conferences and papers in the InfoVis community.
To distinguish between paper, conferences and authors, let us color them:
Select all papers: click
Edit → Findor Ctrl+F, set property: "type", set the filter "= 0".Color the nodes in red: click
Graph Editor → Property, select the 'viewColor' property, checkselected only, clickSet alland choose red.- Repeat for authors (type = 2) and conferences (type = 1) with other colors (here blue and yellow):
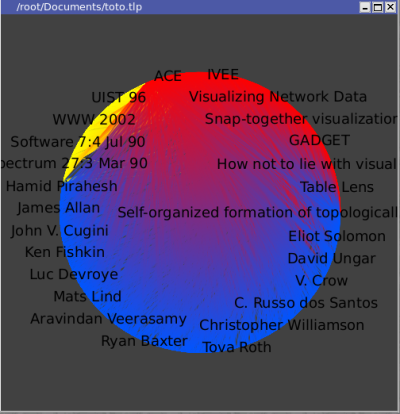
Let's look at a single author, for instance George Robertson.. Select that node with the Find tool, with "titleshort" "=" "G.*Rob.*" (this is a regular expression). Temporarily moving the node away gives a rough estimate of how connected he is:
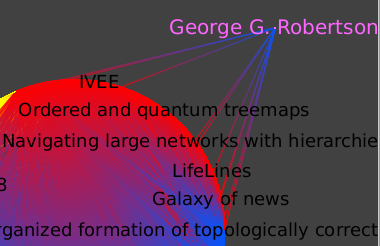
Now, focus on the graph around this author:
- click
Algorithm → Selection → Reachable Sub-Graph, leaving direction to 0 (outgoing edges), startingNodes to viewSelection (nodes selected, in our case simply Mr. Robertson), and distance 1. - Click
Edit → Create Subgraphto save this selection for further manipulation, naming it "GR.1hop.outgoing". Select this new subgraph in the hierarchy tab. We can deselect the nodes now: use selection tool and click away from any node or edge (or Ctrl+Shift+a). - To improve the layout, click
Algorithm → Size → Auto Sizing, then change the layout forAlgorithm → Layout → Hierarchical → Hierarchical Graph. There is an even better hierarchical layout on the plugin server:Help → Plugins → LaBRI Universite Bordeaux 1 → Layout → Sugiyama (OGDF).
We see which papers were published first, as they are cited by the later ones.
Robertson has 11 papers referenced in this database.
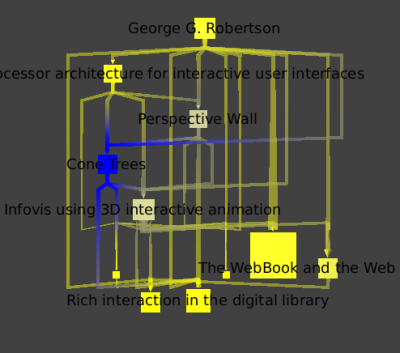
If we apply a coloring by the number of citations (Algorithm → Color → Metric Mapping, with field property set to "arityOut" )
we clearly see that "Cone Trees" is his most influential work:
Now,
- return to the main graph, and make sure the Robertson node is away from the others.
- Select all the papers nodes (
Edit→Find, "property=0"). - We want to add Robertson and all the edges going from or to this node to our selection. To do so, we need the selection tool, and by pressing Ctrl while drawing a rectangle encompassing the node and all of its edges, we add to our current selection.
Save this selection to a new subgraph named GRCitesub:
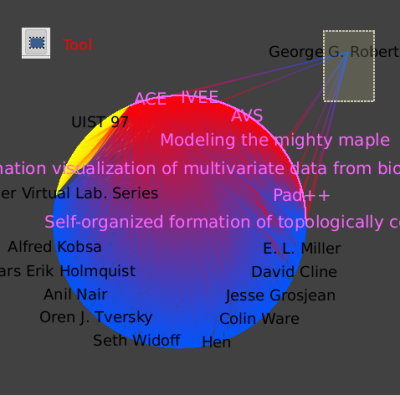
- In the new subgraph, select the Robertson node.
Click
Algorithm → Selection → Reachable Sub-Graphalgorithm again, but with a depth of 2 to find all papers that cite a paper written by Robertson. - Now we want to remove any node or edge that is not selected:
invert the selection (
Edit → Invert Selectionor Ctrl + I), delete (Del). This leaves us with Robertson and all his papers, and the papers citing his papers. - Finally, applying a hierarchical layout yields this picture:

You can now close this graph since we use another one for the next part. The next graph is similar to the this one but, has coauthorship edges linking authors who write papers together. Download it and open it.
Here we focus on the relationship between two authors, in this case Robertson and Card.
-
Select Robertson like before: find, "titleshort" "=" "G.*Rob.*".
Now, add the second one:
Edit → Find, check optionadd, enter the regexp ".*Card.*". -
Move those selected nodes away: use the "selection move" tool
 (if you click on one node, you will be able to move both at the same time):
(if you click on one node, you will be able to move both at the same time):
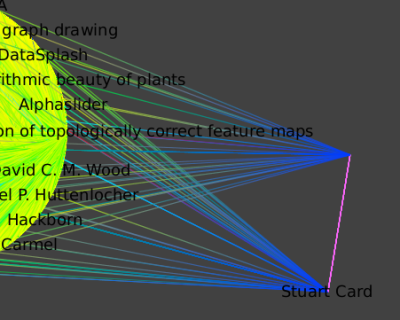
- Select all their neighbors with
Algorithm → Selection → Reachable Sub-Graphwith distance 1. We get the graph of all publications and coauthors of Card and/or Robertson. - Create a new Subgraph, apply a hierarchical layout:

One can repeat those operations for Robertson only and Card only:
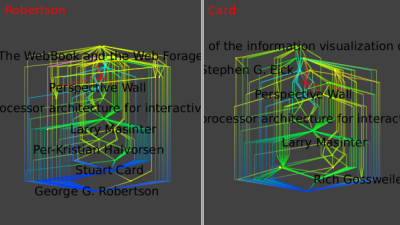
The similarity of these final three images shows the very strong ties between these two authors.
You can now close this graph. The next graph is similar to the old one, but the links are from author to papers, and from papers to conferences. Download it here and open it in tulip.
We used the following to show the large-scale structure of this dataset:- (already done) color authors in green, conferences in blue.
- Consider only the author and conference nodes (i.e. not the paper nodes):
Find "type >= 1",
Algorithm → Selection → Induced subgraph, Create subgraph. - GEM layout (
Algorithm → Layout → Force Directed → GEM)
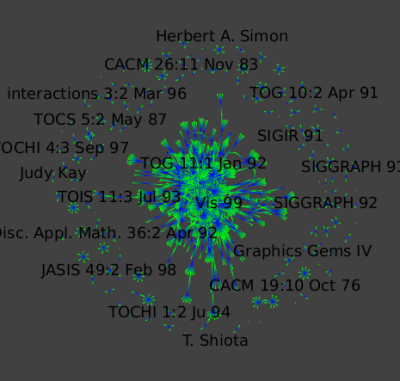
Here is a close-up:
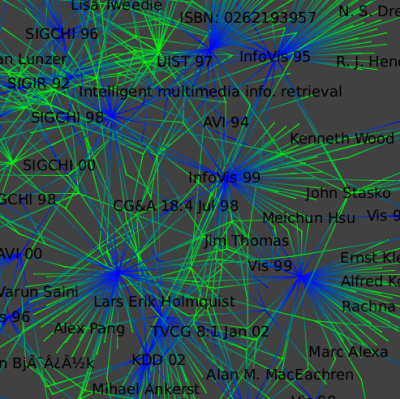
To select only the giant connected component (the largest connected set nodes),
select a node in the center of the giant component and run Algorithm → Selection → Reachable Subgraph
with distance 1000 and directed=2.
- Highlight the structure with
Algorithm → Measure → Graph → Eccentricity, with "Closeness centrality" checked. - To distinguish authors and conferences: select all authors (nodes of type 2) and set their viewLabelColor property to red.
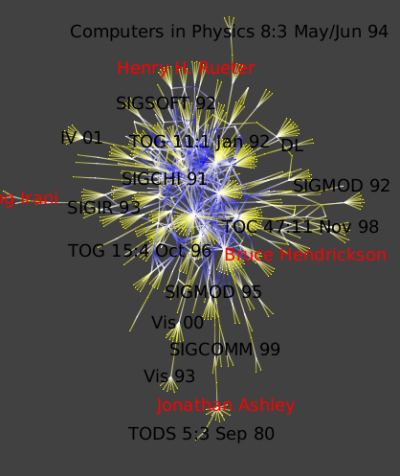
To get a approximation of top conferences, we can look for the ones where the number of authors is high.
To do so, click Algorithm → Measure → Graph → Degree :
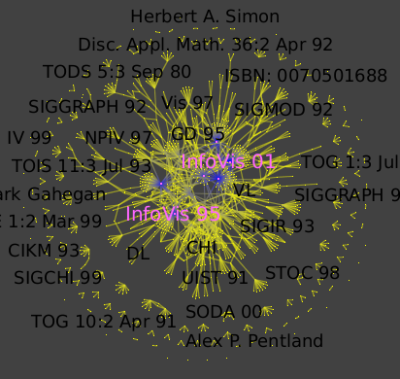
Run Algorithm → Measure → Graph → Strahler to see strong ties between conferences and authors:

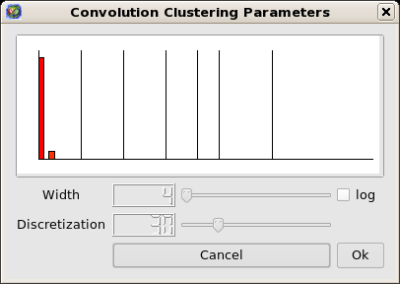
Then run Algorithm → General → Convolution,
the value of the discretization parameter should be near 30 to obtain 5 clusters:
One gets the following clusters:
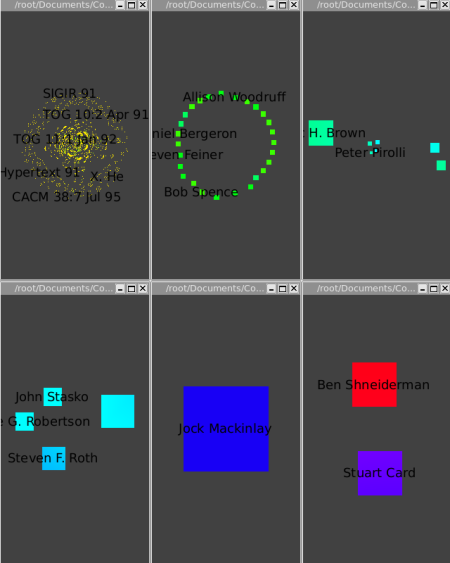
The Strahler-Convolution clustering yields five clusters, according to increasing centrality. The first cluster is mostly yellow, and contains most of the data. The second cluster contains a next tier of 26 authors that have had a relatively strong impact. The third cluster contains a group of 7 influential authors (Chi, Bederson, Eick, Rao, Pirolli, Ward, and Brown), and the fourth cluster (Roth, Robertson, Keim, and Stasko) is yet more central. The fifth cluster is the single node of Mackinlay, and the last one is Card and Shneiderman. Our automatic clustering method clearly yields plausible results in this case.
In the last section, we use this graph.
To see the metanode labels,
Click View editor → Layers Manager, check metanode label.
The clusters have been computed using Algorithm → General → Strength.
The overview image from the previous section, showing the graph of all authors and papers, is extremely cluttered.
The previous section showed one way to extract information (find the most important nodes by convolution clustering).
Another clustering approach, small-world clustering, allows instead to navigate through a hierarchical subdivision of the entire dataset.
The simplified overview allows to understand the graph's high level structure.
The strength metric computes the number of cycles of length 3 and 4 passing through each edge, normalized by the maximum possible value.
The first image shows the clustered dataset.
Small-world navigation is useful when exploring an unfamiliar graph to quickly find the structure of complex components.
The eccentricity metric (Algorithm → Measure → Graph → Eccentricity),
which measures whether nodes are peripheral (here in yellow) or central (here in blue), guides us to complexity immediately.
This metric is O(n^3), but the small-world decomposition simplifies the graph, making the computation tractable.
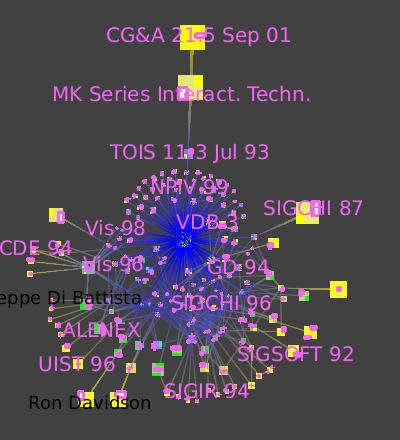
Here is a close-up of the node that has many blue lines leading to it:
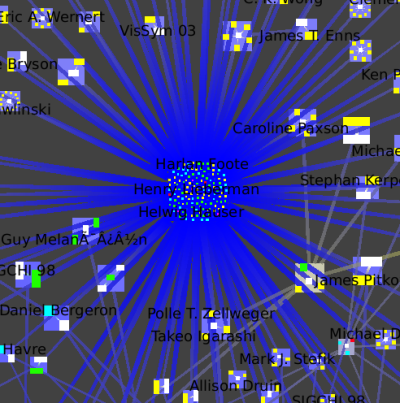
We then open that cluster (right-click on the metanode, go inside), which is itself a small-world graph:

We once again zoom towards the most central node with many blue edges, where we see a cluster containing the InfoVis 96 conference and all the authors who only published at the infovis community in that year.

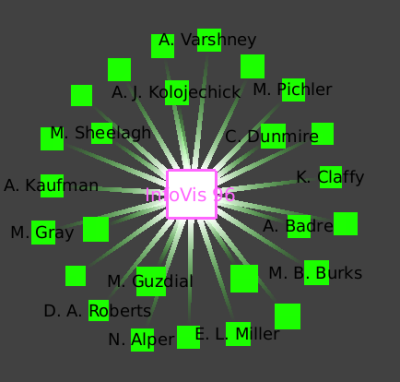
We see this star shaped small-world decomposition only for the InfoVis conferences, because of the nature of this dataset: only the InfoVis conferences have a complete set of authors and papers available.